Dream Challenge: Predicting gene expression using millions of random promoter sequences
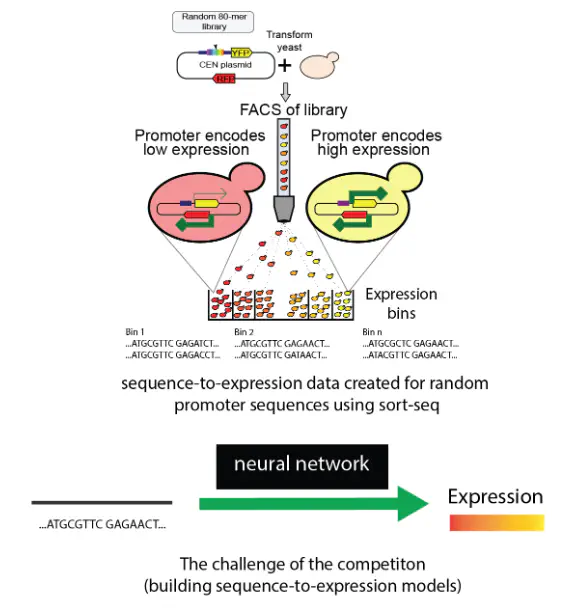
This project is focused on the challenging goal of predicting gene expression values directly from DNA sequences. To achieve this, it employs an end-to-end Graph Attention Network (GAT), a type of neural network that is particularly adept at handling structured data. The project’s approach involves several key aspects:
- Utilizing DNA Sequences: The primary input for the model is DNA sequence data. These sequences are inherently complex and contain the genetic information that ultimately influences gene expression.
- Graph Attention Network: The GAT model is chosen for its ability to process data in a graph structure, which is well-suited to representing the interconnections and relationships within genetic data. The attention mechanism in GATs allows the model to focus on specific parts of the graph (i.e., certain regions of the DNA sequence) that are more relevant for predicting gene expression levels.
- End-to-End Learning: The model is designed to be end-to-end, meaning it takes raw DNA sequences as input and directly outputs predicted expression values. This approach eliminates the need for manual feature extraction, allowing the network to learn the most relevant features for prediction by itself.
- Predicting Expression Values: The ultimate goal is to accurately predict the level of gene expression based on the given DNA sequence. This has significant implications for understanding genetic regulation and can be applied in various fields, including personalized medicine and genetic research.
By leveraging the capabilities of GATs, the project aims to provide a more nuanced and accurate understanding of how specific sequences within DNA contribute to gene expression, potentially leading to breakthroughs in genomics and related areas.